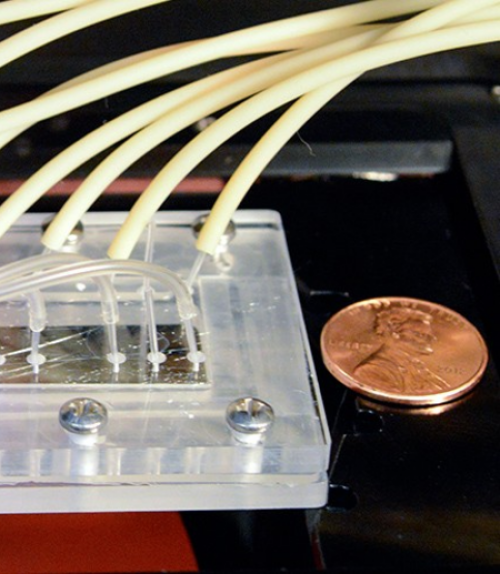
Cancer cells are a wily adversary. One reason the disease outfoxes many potential treatments is because of the diversity of the cancer cell population. Researchers have found this population difficult to characterize and quantify.
A Cornell-led team took a novel, interdisciplinary approach to analyzing the behavior of breast tumor cells by employing a statistical modeling technique more commonly used in physics and economics. The team was able to demonstrate how the diversity, or heterogeneity, of cancer cells can be influenced by their chemical environment – namely, by interactions with a specific protein, which leads to tumor growth.
The researchers’ paper, “Lymphoidal Chemokine CCL19 Promoted the Heterogeneity of the Breast Tumor Cell Motility Within a 3D Microenvironment Revealed by a Lévy Distribution Analysis,” published Feb. 14 in Integrative Biology.
“It’s pretty tough to treat cancer. A lot of people in the field believe that is because of the diversity in the cancer population,” said senior author Mingming Wu, professor of biological and environmental engineering in the College of Agriculture and Life Sciences. “While immune cells are rounded and kind of similar and move in the same way, cancer cells are different in shape and move at different speed. We know that fast movers are very lethal. How would you quantify that heterogeneity?”
Another challenge is that only a few cancer cells move fast and do the most damage, and they’re difficult to find.
The effort to track these rare cells is similar to the search for elusive particles being conducted in the lab of co-author Anders Ryd, professor of physics in the College of Arts and Sciences. During a conversation over coffee, Ryd and Wu realized the cancer cell research could incorporate the same type of sophisticated statistical tools that have helped particle physicists understand rare energy phenomena, such as the much-sought Higgs boson.
“What we do in particle physics is really statistical data analysis, trying to figure out what functional forms describe our data,” Ryd said. “And in this case here, the interest was to look at the outliers, the cells that migrated further, and characterize that. A lot of the tools that we are using in particle physics lend themselves very well to this analysis.”
Wu’s Biofluidics Lab worked with the Cornell NanoScale Science and Technology Facility (CNF) to fabricate a microfluidic device with three parallel channels, each roughly the width of a human hair. The team introduced breast tumor cells into the device, along with the chemokine protein CCL19, which is secreted by lymph nodes and is highly expressed in malignant tumor cells.
In order to model the cancer cell trajectories, the team used Root, an open-source software for performing statistical analysis in high-energy physics and in certain economics applications.
The researchers found that the presence of chemokine caused the targeted cancer cells to move faster, and heterogeneity increased.
“It is similar to how we as a society are trying to make the population more diverse, because we know that if the population is diverse, it’s more robust, more healthy,” Wu said. “I think that cancer is the same way. They are making their population more diverse, more indestructible.”
A treatment that inhibits the receptor to CCL19 could potentially decrease the invasiveness of tumor cells, although that might also cause the cancer cells to adopt new, even stealthier strategies to survive, Wu said.
By analyzing how these cells respond to environmental cues – such as chemical gradients, temperature, light intensity and mechanical force – Wu’s team hopes to elucidate the underlying principles of biology, which aren’t as cut and dried as the fundamental laws of physics.
Her team may borrow a few techniques, too.
“There are a lot of tools in physical science already, because physical science has always been a very quantitative field,” Wu said. “It’s only recently that quantitative biology is starting to shape up. So I feel like this integration is powerful because you don’t have to reinvent the wheel to do this modeling.”
Other contributors included co-lead authors Beum Jun Kim, Ph.D. ’04, now a senior engineer at Rheonix, and Pimkhuan Hannanta-anan ’12, a faculty member at King Mongkut’s Institute of Technology Ladkrabang in Thailand; and Melody Swartz, professor of molecular engineering at the University of Chicago.
The research was supported by the National Cancer Institute, the Swiss National Science Foundation, the Cornell Center on the Microenvironment and Metastasis, the Cornell Nanobiotechnology Center and CNF.