Combining state-of-the-art X-ray technology and cryogenics, Cornell physics researchers have developed a new method for analyzing proteins in action, a breakthrough that will enable the study of far more proteins than is possible with current methods.
Jonathan Clinger, a postdoctoral researcher in the Department of Physics, in the College of Arts and Sciences (A&S), is lead author of the study, which was published Aug. 4 in the journal of the International Union of Crystallography.
Current X-ray crystallography methods involve making “movies” – actually a series of snapshots – of a protein’s atomic structure over time, to reveal the steps in an enzymatic reaction, or how a photoactive protein responds to light. These images inform the design of optimized enzymes for biotechnology and pharmaceuticals.
The most powerful current methods involve triggering a reaction within a protein crystal, collecting an X-ray diffraction pattern after a short time delay, and then repeating with different time delays. X-ray data from hundreds to tens of thousands of small protein crystals must be collected and processed to obtain a three-dimensional snapshot of the protein structure at just one time delay.
“These are all very bespoke experiments that require a lot of overhead,” Clinger said.
These methods require a complex experimental setup at a synchrotron or X-ray free-electron laser source, a team of scientists to operate it, massive amounts of protein and complex data analysis, making them largely inaccessible to most researchers. Only a handful of protein systems have been studied so far.
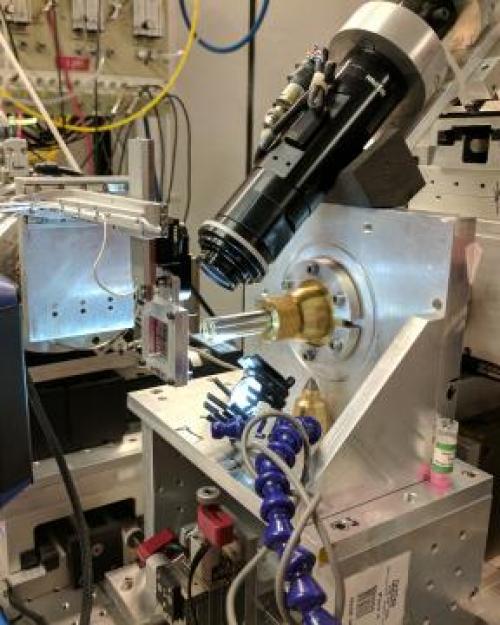
In the new method – millisecond mix-and-quench crystallography, or MMQX, developed at the Cornell High Energy Synchrotron Source (CHESS) – a reaction is triggered by rapidly applying a reactant-containing solution to a protein crystal. The reaction is then allowed to proceed for a desired time.
But instead of trying to quickly collect X-ray data before the reaction proceeds further, the crystal is cooled in a few milliseconds to 100 kelvins (minus 279.67 degrees Fahrenheit). The reaction is quenched – literally stopped cold – so X-ray data can be collected later.
“Our approach allows anyone to get time-resolved data, using standard synchrotron beamlines and workflows,” said senior author Robert Thorne, professor of physics (A&S) and a Weiss Presidential Fellow.
Far more data per crystal can be collected, Thorne said, in part because at cryogenic temperatures, protein crystals can tolerate much larger X-ray doses before showing damage. Data can be collected by one or more scientists working remotely using mail-in protocols on any automated crystallography beamline, at any synchrotron source.
And now a single crystal can be sufficient, rather than hundreds of thousands, to obtain a snapshot of the protein structure at a given time delay.
A key to the method is technology for ultra-rapid cooling of protein crystals based on earlier discoveries in the Thorne lab and commercially developed by Thorne’s company, MiTeGen. Postdoctoral researcher David Moreau, Ph.D. ’20, and Clinger constructed a mix-and-quench instrument and collaborated with Matt McLeod and Todd Holyoak of the University of Waterloo to study the enzymatic mechanism of PEPCK, an essential enzyme in glucose metabolism.
The 40-millisecond time resolution in these proof-of-concept experiments is comparable to what has been achieved with other methods that require three to five orders of magnitude more crystals. And, according to Thorne, 1- to 2-millisecond time resolutions are feasible.
“Time-resolved atomic resolution structural study of proteins need no longer be a specialist pursuit,” Thorne said. “MMQX will make it routine. It can be a powerful addition to the toolkit used by biologists and biotechnologist to dissect, optimize and inhibit biomolecular function.”
The Center for High Energy X-ray Sciences (CHEXS) is funded by the National Science Foundation; the Macromolecular Diffraction at CHESS (MacCHESS) is funded by the National Institutes of Health and by the state of New York.
Susan Newman is a science communicator for the Cornell Laboratory for Accelerator-based Sciences and Education.